
Predicting how a potential drug behaves inside the body is one of the most critical challenges in early drug development. Among the key properties, cell permeability determines how well a compound can cross biological barriers and reach its intended target.
To support faster and more accurate decision-making, SMAG has developed a new MDCK (Madin-Darby Canine Kidney) cell permeability prediction model - a powerful addition to our TOPIA-ADMET suite.
This machine learning-based model allows researchers to predict permeability in silico with high precision, reducing reliance on costly and time-consuming experimental assays.
Why MDCK Permeability Prediction Matters
The MDCK cell line is widely accepted as an in vitro model for evaluating drug permeability. It is especially important for:
- Passive diffusion studies - understanding how compounds move across epithelial barriers.
- Efflux transporter activity - assessing mechanisms critical for central nervous system (CNS)-targeted drugs.
- Oral absorption evaluation - determining whether orally administered compounds can effectively enter circulation.
Traditional lab methods such as permeability assays are accurate but not always scalable during early-stage research. By introducing computational predictions, researchers can prioritize compounds earlier and allocate resources more efficiently.
Inside SMAG’s MDCK Permeability Model
Our in-house data science and pharmacology teams designed this model to be both robust and practical.
Model Highlights
- Input Features: Morgan fingerprints generated from SMILES strings
- Algorithm: Random Forest Regression
- Output: Continuous prediction of log Papp (nm/s) values
- Use Case: High-throughput screening of novel compounds without complex descriptors
This design ensures the model is simple to use, scalable, and easily integrated into existing drug discovery pipelines.
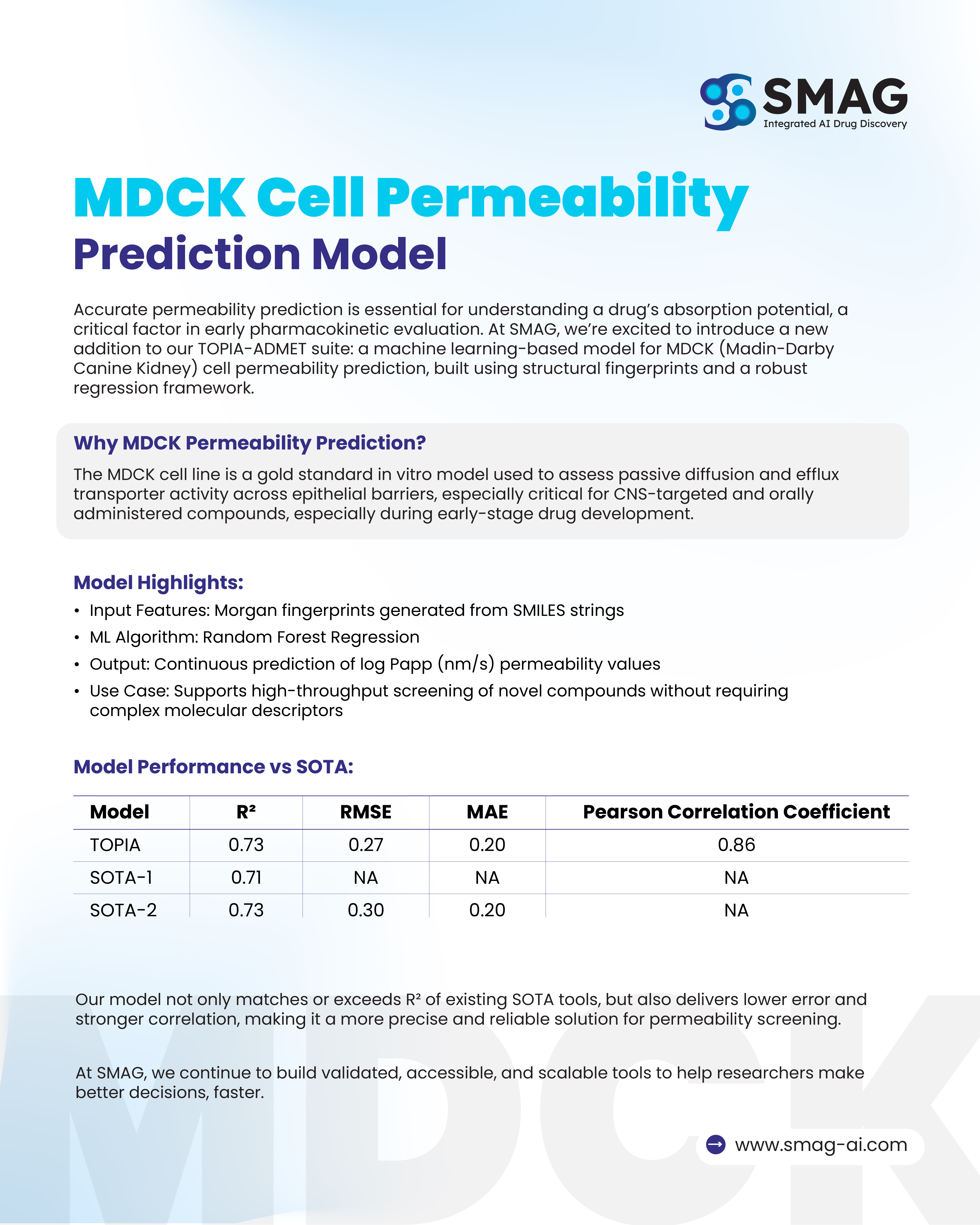
Benchmarking and Performance
Our model has been rigorously validated against state-of-the-art (SOTA) tools.
Performance Metrics: R² (Coefficient of Determination), RMSE (Root Mean Squared Error), MAE (Mean Absolute Error), Pearson Correlation
| Model | R² | RMSE | MAE | Pearson Correlation |
|---|---|---|---|---|
| TOPIA | 0.73 | 0.27 | 0.20 | 0.86 |
| SOTA-1 | 0.71 | NA | NA | NA |
| SOTA-2 | 0.73 | 0.30 | 0.20 | NA |
Key Takeaways:
- Achieves equal or higher accuracy than comparable models.
- Produces lower error (RMSE), improving prediction reliability.
- Demonstrates strong correlation (0.86), ensuring confidence in screening results.
Applications in Drug Discovery
The MDCK permeability prediction model is designed for real-world applications, including:
- Lead optimization: Filter and prioritize promising drug candidates.
- CNS research:Assess the potential for compounds to cross brain-related barriers.
- ADMET profiling Integrate with other TOPIA-ADMET models for a comprehensive pharmacokinetic view.
- Preclinical strategy:Reduce unnecessary wet-lab testing by using in silico screening first.
Looking Ahead
At SMAG, our focus is on developing validated, accessible, and scalable tools that help the scientific community make faster, data-driven decisions. The MDCK permeability model reflects this mission by combining scientific rigor with computational efficiency.
It is the latest advancement within our TOPIA-ADMET suite, and it marks another step toward transforming how early-stage research is conducted.
Frequently Asked Questions (FAQs)
Conclusion
The new MDCK permeability prediction model underscores SMAG’s commitment to building innovative, research-driven solutions that accelerate the pace of discovery. By integrating this tool into early-stage workflows, researchers can screen compounds more effectively, cut down experimental costs, and make better-informed decisions.
Interested in learning more or collaborating? Contact us now